What’s Your CC7? - Sat, 28 Sep 2024
If you’re not a recent user of WikiTree, you might not be familiar with the term CC7. It stands for Connection Count at 7 Degrees.
One “Degree” at WikiTree is defined to be a connection to a parent, sibling, child or spouse. Your connections up to 7 degrees are your shortest path to any other person.
Therefore your CC7 will include your descendants and everyone up to your great-great-great-great-great grandparents, your great-great-great-great grand uncles and aunts, your first cousins 3 times removed, your 2nd cousins twice removed, and your 3rd cousins once removed. These would be all your relatives that are included in your CC7.
But the interesting catch is the inclusion of a spouse as a CC7 connection. This allows spouses of your descendants, spouses of your great-great great grand uncles and aunts, spouses of your first cousins twice removed, spouses of your second cousins once removed and spouses of your third cousins, and your spouses as well.
If the spouse is not already at 7 degrees, then you can include their relatives as well, and even spouses of their relatives and relatives of spouses of their relatives. Whew it gets complicated.
But Why Spouses?
Some genealogy software have a feature that tells you the shortest path between two people. The idea was popularized by the Bacon Number, that any Hollywood actor connects to actor Kevin Bacon through at most 6 degrees of separation, where a connection is a film role. The idea was made possible in genealogy software when computers became sufficiently powerful to be able to go through the billions of possible connections in a short amount of time to find the closest connection.
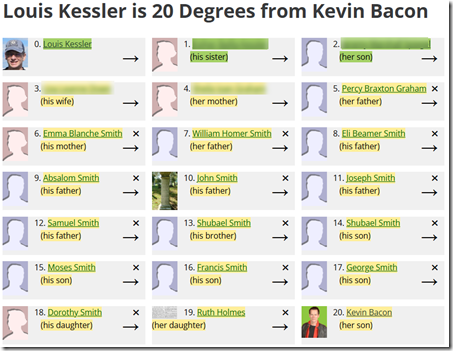
Well, I don’t have a Bacon number because I’ve never acted in any Hollywood films, but just for fun I can find my WikiTree connection count with Kevin Bacon:
It turns out (and I just discovered this when I did it) that my niece is a seventh cousin twice removed with Kevin Bacon!
I was going to comment about all the husband and wife connections to make this possible, but in this case there was only one, the connection between my nephew and my niece.
Spouses could have been sprinkled throughout that connection, and you could imagine how with a sufficiently large one-world tree like at WikiTree you can connect to almost everybody. It’s just a matter of how many degrees.
But if spouses weren’t included, you’d be limited to only blood relatives.
Wiki Tree 2024 Challenges
What raised my awareness of the CC7 was the current set of Challenges taking place this year at WikiTree. Every few weeks, WikiTree would take a well-known genealogist, and WikiTree members would work together to expand that genealogist’s CC7 on WikiTree.
So far this year the subject genealogists have been:
- Meli Alexander (CC7 increased from 1378 to 2227)
- Lianne Kruger (564 to 1368)
- Randy Seaver (1621 to 2457)
- Judy Muhn (795 to 1498)
- David Allen Lambert (2290 to 3338)
- Ellen Thompson-Jennings (1170 to 1834)
- Thomas MacEntee (1039 to 1524)
- Lisa Louise Cooke (935 to 1360)
- Jill Ball (2154 to 3022)
- Melanie McComb (1290 to 1851)
- Jarrett Ross (547 to 1498)
So for a full week, about 30 enthusiastic WikiTree volunteers managed to add about 800 CC7 profiles for each genealogist, while at the same time adding sources and breaking a few of their brick walls
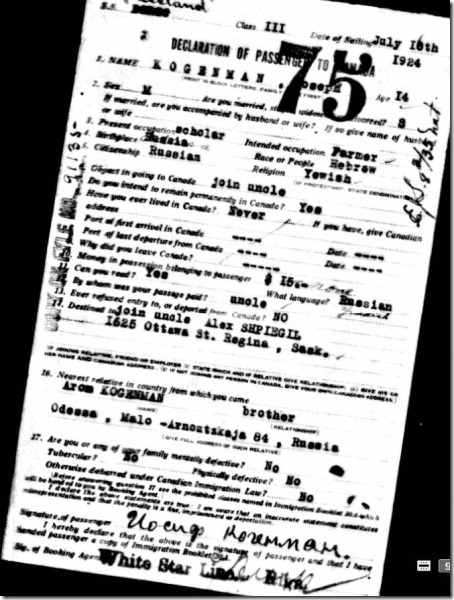
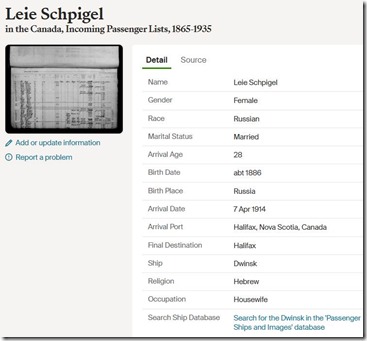
I was interested in both Melanie’s and Jarrett’s Jewish connections, so I become a volunteer in both their weeks and helped add to their profiles and shared knowledge and learned a lot myself on the group’s challenge Discord channel. You can see Melanie’s “Final Reveal” on YouTube where I was one of the panelists. And you can watch Jarrett’s “Final Reveal” which will take place on October 3.
The Importance of CC7
Many people likely have heard of the FAN Principle, which is often promoted by Elizabeth Shown Mills and others. FAN stands for Friends, Associates and Neighbors, and the idea is that in order to learn more about your relatives, you will find clues and records if you look a their FAN Club.
Like many other genealogists, I concentrate primarily on my own relatives (through blood or adoption) and will include their spouses and stepfamily. But I do stop there and don’t go into each of my relatives’ spouses families – their ancestors, siblings, cousins, etc.
Well a CC7 though marriage is very similar to studying a FAN club. Many of these cousins through marriage lived in the same places, associated with, and may hold clues that lead to records of your direct relatives. At a limit of 7 degrees, we usually are staying close enough to the relative that these people are still quite relevant. In addition, any family trees that include some of these relatives through marriage will overlap with your tree and include that relative’s descendants and possibly have better info on them than you have.
What is also fun that you can do with a CC7 at WikiTree is find the notable people you are connected to. During Jarrett’s challenge, we found he was directly related within 7 degrees to four “Notables” and was a CC7 to another 16 relatives through marriage (including Cary Grant!).
I still don’t plan to add relatives through marriage to my primary tree on MyHeritage. But I do plan to use WikiTree to keep track of my CC7s for me. My CC7 count at WikiTree is currently only 756 and I have no people marked “Notable” yet among them. I plan to spend a bit of time each week to expand my CC7 connections there. It really is interesting and sometimes helpful as well to look at your relatives’ relatives.
In doing so, I may find some CC7s that are already in the main tree. Connecting them will shorten some of my paths to the notables and some might land within 7 degrees. Maybe I can even reduce my connection to Kevin Bacon down from 20.




![clip_image002[12] clip_image002[12]](https://www.beholdgenealogy.com/blog/wp-content/uploads/Creating-a-New-Tree-on-MyHeritage_9C8C/clip_image00212_thumb.jpg)
 Feedspot 100 Best Genealogy Blogs
Feedspot 100 Best Genealogy Blogs





