In my last post, I was able to create a raw data file from the Whole Genome Sequencing (WGS) BAM file using the WGS Extract program. It seemed to work quite well.
But my previous post to that: WGS – The Raw VCF file and the gVCF file, I was trying to see if I could create a raw data file from the Variant Call Format (VCF) file. I ended that post with a procedure that I thought could generate a raw data file, which was:
- Make a list of all the SNPs you want raw data for.
- Initially assign them all the human genome reference values. Note: none of the VCF files give all of these to you, so you need this initially set this up. Wilhelm HO has good set of them included with his DNA Kit Studio.
- The positions of variants in your gVCF file should be marked as no-calls. Many of these variants are false, but we don’t want them to break a match.
- The positions of variants in your filtered VCF should be marked as having that variant. This will overwrite most of the optimistic no-calls marked in step 3 with filtered reliable values.
When I wrote that, I had thought that the gVCF file contained more variants in it than the Raw VCF file had. During my analysis since then, I found out that is not true. The Raw VCF contains all the unfiltered variants. Everything that might be considered to be a variant is in the Raw VCF file. The gVCF includes the same SNP variants that are in the Raw VCF file, but also includes all the insertion/deletions as well as about 10% of the non-variant positions. It’s the non-variant positions that makes the gVCF such a large file.
So right away, in Step 3 of the above proposed procedure, the Raw VCF file can be suggested instead of the gVCF file and will give the same results. That is a good thing since the Raw VCF file is much smaller than the gVCF file so it will be faster to process. Also the Raw VCF file and the filtered VCF file include the same fields. My gVCF included different fields and would need to be processed differently than the other two.
(An aside: I also found out that my gVCF supplied to me by Dante did not have enough information in it to determine what the variant is. It gives the REF and ALT field values, but does not include the AC field. The AC field gives the frequency of the ALT value, either 1 or 2.
- If REF=A, ALT=C, AC=1, then the variant value is AC.
- If REF=A, ALT=C, AC=2, then the variant value is CC
- If REF=A, ALT=C, AC is not given, then the variant value can be AC or CC.
For me to make any use of my gVCF file, for not just this purpose but any purpose, I would have to go back and ask Dante to recreate it for me and include the AC field in the variant records. End aside.)
Estimating Type 1 and Type 2 Errors
We now need to see if the above procedure using the Raw VCF file in step 3 and the filtered VCF file in step 4 will be accurate enough to use.
We are dealing with two types of errors.
Type 1: False Positive: The SNP is not a variant, but the VCF file specifies that it is a variant.
Type 2: True Negative: The SNP is a variant, but the VCF file specifies that it is not a variant.
Both are errors that we want to minimize, since either error will give us an incorrect value.
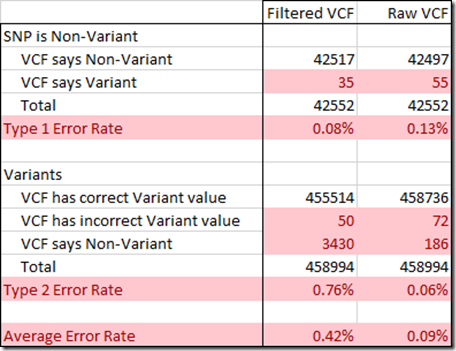
To determine the Type 1 and Type 2 error rate, I used the 959,368 SNPs that the WGS Extract program produced for me from my BAM file. That program uses well a developed and respected genomic library of analysis functions called samtools, so the values it extracted from my WGS via my BAM file are as good as they can get. It is essential that I have as correct values as possible for this analysis, so I removed 2,305 values that might be wrong because some of my chip test results disagreed with. I also removed 477 values that WGS Extract included but were at insertion or deletion positions.
From the remaining values, I could only use positions where I could determine the reference value. This included 458,894 variant positions, which always state the reference value, as well as the 10% or so of non-variant reference values that I could determine from my gVCF file. That amounted to 42,552 non-variants.
Assuming these variant and non-variant positions all have accurate values from the WGS extract, we can now compute the two types of errors for my filtered VCF file and for my Raw VCF file.
In creating a VCF, the filtering is designed to eliminate as many Type 1 errors as possible, so that the variants you are given are almost surely true variants. The Raw VCF only had 0.13% Type 1 errors, and the filtering reduced this to a very small 0.08%.
Type 1 and Type 2 errors work against each other. Doing anything to decrease the number of Type 1 errors will increase the number of Type 2 errors and vise versa.
The Raw Data file turns out to only have 0.06% Type 2 errors, quite an acceptable percentage. But this gets increased by the filtering to a whopping 0.76%.
This value of 0.76% represents the number of true variants that are left out of the filtered VCF file. This is what is causing the problem with using the filtered VCF file to produce a raw data file. When the SNPs that are not in the filtered VCF file are replaced by reference values, they will be wrong. These extra errors are enough to cause some matching segments to no longer match. And a comparison of a person’s raw dna with his raw dna generated from a filtered VCF file will not match well enough.
If instead, the Raw VCF file is used, the Type 2 errors are considerably reduced. The Type 1 errors are only slightly increased, well under worrisome levels.
Since there are approximately the same number of variants as non-variants among our SNPs, the two error rates can be averaged to give you an idea of the percentage of SNPs expected to have an erroneous value. Using the Raw VCF instead of the filtered VCF will reduce the overall error rate down from 0.42% to 0.09%, a 79% reduction in errors.
This could be reduced a tiny bit more. If the Raw VCF non-variants are all marked as no-calls, and then the Filtered VCF non-variants are replaced by the reference values, then 20 of the 55 Type 1 Errors in my example above, instead of being wrong, will be marked as no-calls. No-calls are not really correct, but they aren’t wrong either. For the sake of reducing the average error rate from 0.09% to 0.07%, it’s likely not worth the extra effort of processing both VCF files.
Conclusion
Taking all the above account, my final suggested procedure to create a raw data file from a VCF file is to use only the Raw VCF file and not the filtered VCF file, as follows:
- Make a list of all the SNPs you want raw data for.
- Initially assign them all the human genome reference values. Note: none of the VCF files give all of these to you, so you need this initially set this up. Wilhelm HO has good set of them included with his DNA Kit Studio.
- Mark the positions of the variants in your Raw VCF with the value of that variant. These will overwrite the reference values assigned in step 2.
Voila! So from a Raw VCF file, use this procedure. Do not use a filtered VCF file.
If you have a BAM file, use WGS Extract from yesterday’s post.
Update: May 14: Ann Turner pointed out to me (in relation to my “Aside” above, that in addition to the AC (allele count) field, the GT (genotype) field could supply the information to correctly identify what the variant is. Unfortunately, the gVCF file Dante supplied me with has missing values for that field’s values.
I’ve looked at all the other fields in my gVCF file and entries that leave out the BaseQRandSum and ClippingRankSum fields as they often indicate a homozygous variant, but I’ve found several thousand SNPs among the variants that constitute too many exceptions to use this as a "rule".
Wilhelm HO is working on implementing the sort of procedure I suggest into his DNA Kit Studio. It likely will be included when he releases Version 2.4, and his tool will then be able to produce a raw data file from a VCF file and will also extract a mtDNA file for you that you can upload to James Lick’s site for mtDNA Haplogroup analysis.