A couple of months ago, I compared my VCF file to my DNA test results.
The Variant Call Format (VCF) file is given to you when you do a Whole Genome Sequence (WGS) test. That test finds your DNA values for your whole genome, all 3 billion positions, not just the 700,000 or so positions that a standard DNA test gives you.
But most of those 3 billion positions are the same for most humans. The ones that differ are called Single-Nucleotide Polymorphisms (SNPs) because they “morph” and can have differing values among humans. The standard DNA companies test a selection of the SNPs that differ the most, and they can use the 700,000 they selected for matching people without having to test all 3 billion positions. It works very well. WGS tests are not needed for finding relatives.
Converting VCF to a Raw Data File
But near the end of my last post, I was trying to see if the VCF file could be converted into a raw data file that could be uploaded to GEDmatch or a DNA company that allows raw data uploads.
My VCF file contains 3,442,712 SNPs whose value differ for me from the standard human reference genome. Of those, I found 471,923 SNPs were the same SNPs (by chromosome and position) as those in my raw data file that I created by combining the raw data from 5 companies (FTDNA, MyHeritage, Ancestry, 23andMe and LivingDNA). I compared them in my first analysis and found that 2,798 of them differed, which is only 0.6%.
At the time, I didn’t think that was too bad an error rate. So I thought a good way to make a raw data file from a VCF file would be:
- Take a raw data file you already have to use as a template.
- Blank out all the values
- Add values for the positions that are in the VCF file
- Fill in the others with the human reference genome value.
The basis of that idea is that if it’s not a variant in the variant file, then it must be the reference value.
Today on Facebook, Ann Turner told me that that’s not necessarily the case. The reason she believes, is that the VCF file does not contain all the variant SNPs. And the discrepancies were enough to break her comparison of “herself” with “herself” into 161 segments.
So What’s Really Different Between VCF and Raw Data?
In my first analysis, I only compared whether the values were the same or not, giving that 0.6% difference. I did not look at the specific values. Let’s do that now:
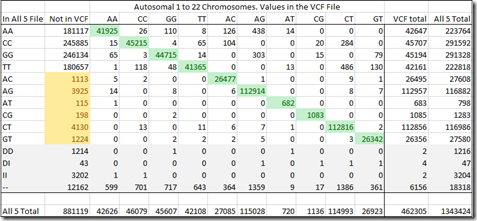
For this analysis, let’s not worry about the rows: DD (Deletions), DI (Deletion/Insertions), II (Insertions) or – (no-calls), since they are only in the raw data and not in the VCF file.
The green values down the diagonal are the agreement between the All-5 raw data file and the VCF file. Any numbers above and below that diagonal are disagreements between the two. Those are the 0.6% where one is wrong for sure, but we don’t know which.
But let me now point you to those yellowed numbers in the “Not in VCF” column. Those are all heterozygous values, with two different letters. AC, AG, AT, CG, CT or GT. If they have two different letters, then they cannot be human reference values. One of the two letters is a variant and those entries should have been in the VCF file. But they were not.
This creates even a bigger concern than our earlier 0.6% mismatch. If we total these yellow counts, we find there’s 10,705 or 1.2% of the 881,119 SNPs that are not in the VCF file that should have been in the VCF file.
Again, we don’t know who is wrong, the raw data, or the VCF file. But from Ann’s observations, we’d have to say at least some of those heterozygous values must have been left out and when reference values were added instead to Ann’s file, they caused the match breaking that resulted in 161 segments.
Which is Correct: VCF or Raw Data
When you are comparing two things, and you know one is wrong, you don’t know which of the two is the wrong one. You need others to compare with. I am awaiting the results of my long read WGS test, and when that comes I’ll have a third to compare.
But until then, can I get an idea of which of the two files might be more often correct? There’s one thing I can do.
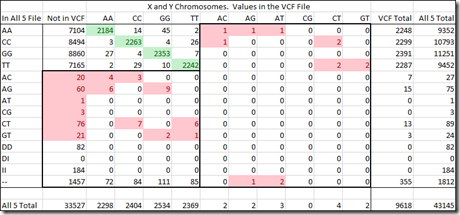
I can provide the same table as I did above, but for the X and Y chromosomes. Since I’m a male, I only have one X and one Y chromosome. The value could be shown as a single value, but it is still read as a double value and therefore shown as a double value. The caveat is that the two letters must be the same or the read is definitely incorrect. (Note that this table excludes 688 SNPs that are in the pseudoautosomal region of the X or Y which can recombine and have two different allele values).
So let’s see the table:
The top left section contains the agreed upon values (in green) between the All 5 raw data file and the VCF file. The counts in that section above and below the green values are disagreements between All 5 and VCF and we don’t know which is correct and which is wrong.
The numbers in red are incorrect reads. Those on the left side are incorrect reads for the All 5 raw data file. It has 219 incorrect reads versus 40,848 correct reads, a ratio of 1 every 186.
The right side are incorrect reads for the VCF file. It has 13 incorrect reads versus 9,605 correct reads. That’s a ratio of 1 every 739 reads.
Now, verging on the realm of hyperbole, the difference in ratios could indicate that an error in a standard DNA test is 4 times (739 / 186) more common than an error in a VCF file.
And applying that ratio to the 10,705 heterozygous values that should have been in the VCF file, we would say that 8,564 would be because the raw data file is wrong, and 2,141 because the VCF file should have included them but did not.
And if 2,141 values out of your DNA file created from the VCF file are incorrect, couldn’t that quite easily have caused the 161 segments that Ann observed?
Yes, this is all conjecture. But the point is that maybe the VCF file is leaving out a significant number of variants. If that is the case, then we can’t just put in a reference value when there is no value in a VCF file. And that would mean a raw data file created from a VCF file and filled in by human reference values may not have enough accuracy to be usable for matching purposes.