My last post defined base pairs and centimorgans, explained their relationship with each other, checked the accuracy of one genetic map, and described 3 converters that will calculate cM from base pairs.
Before leaving this topic, I wanted to document what I tried in an attempt to create an accurate bp to cM map using segment match files.
Segment Match Files
Segment Match Files contain all the matches for a person. You can download them from Family Tree DNA, 23andMe, MyHeritage DNA or GEDmatch-Tier1.
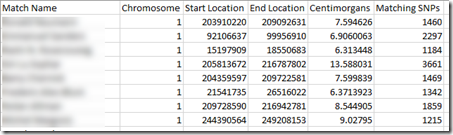
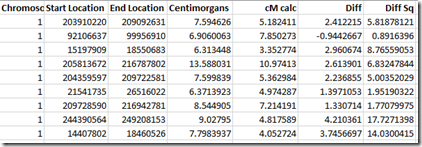
For each segment match, they provide at least the name of the person you match with as well as the chromosome, starting and ending base pair, cM, and number of SNPs. Here is an example of the beginning of a segment match file from Family Tree DNA:
These Family Tree DNA’s bp to cM map with the Centimorgan value shown with lots of decimal places. It says, for example that that the segment on chromosome 1 from bp 203,910,220 to bp 209,092,631 is 7.594626 cM.
There are also a lot of segments given in Family Tree DNA’s segment match files. My file lists 188,438 segments.for the 32,449 people that I match to.
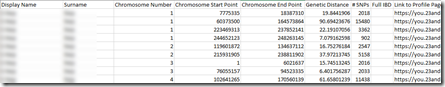
23andMe’s segment match file looks like this:
and it has more information to the right about the person matched to. It also gives an accurate cM value (e.g. 19.8441906) which it called the “Genetic Distance”
However, I only have 10,828 segments in my match file because 23andMe limits to 1500 people, which can be increased to 5,000 with a subscription to their Plus service.
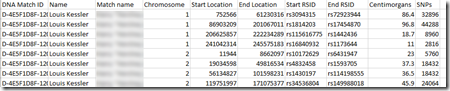
MyHeritage DNA’s segment match file looks like this:
They do not have an accurate cM value. It is fine for most purposes but is rounded off to a tenth of of cM, e.g. 86.4.
My MyHeritage file has 75,028 segments for 19,162 people.
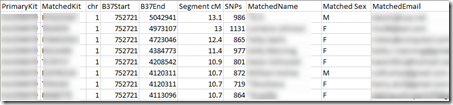
Finally, GEDmatch’s segment match file looks like this:
Like MyHeritage, GEDmatch also rounds their cM values to tenths of a cM.
My GEDmatch file only has 10,000 segments which is the limit GEDmatch allows. Those are for 1,955 people.
What is the Goal?
I want to come out with a map that for a particular company, will map a bp position to a cM genomic position on each chromosome. Then if you have the bp at the start of a segment and the bp at the end of the segment, you can determine the genomic positions at the start and the end of the segment. The cM of the segment then can be determined by subtracting the starting genomic position from the ending genomic position.
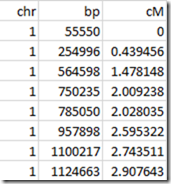
So we want a table that looks like:
This table is from the one that Amy Williams and Jonny Perl use.
So if we have a segment from 564,598 bp to 1,100,217 bp, then that segment would be 2.743511 – 1.478148 = 1.265363 cM.
If we had a start or end position in between two of those values, then would could interpolate. e.g.: at bp = 850,000, the cM would be:
cM = 2.028035
+ (2.595322 – 2.028035) * (850,000 – 785,050) / (957,898 – 785050)
which equals 2.241201 cM.
This system works well when the programmer is using a database which has a fast lookup for entries on either side of the lookup value 850,000.
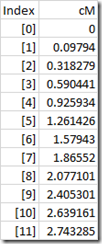
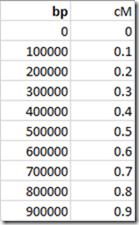
Alternatively, this can be approximated and simplified by interpolating values every 100,000 bp and setting them up in a simple array:
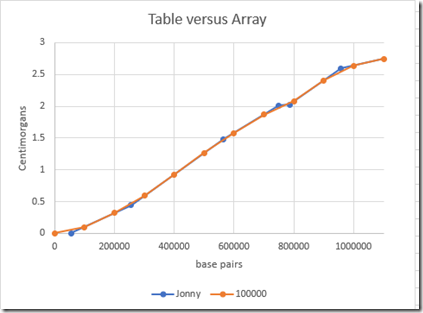
These are now interpolated and no longer exact. Here’s a comparison of the Table values versus the Array values:
You can barely see the differences betwen the two. So the array values should be good enough to get segment cM within 0.1 cM.
The advantage of storing this in an array is that it simplifies programming, uses less memory and is faster to look up and calculate. With bp = 850,000, we know without lookup to use the [8] and [9] entries, and the interpolation becomes:
cM = 2.077101 + (2.405301 – 2.077101) * (850,000 – 800,000) / 100,000
equalling 2.241201 cM
which in this case happens to be exactly what the result was for the array method. That’s only because there are no array points between 700,000 and 800,000. If there were, the results would slightly differ.
Okay. That’s what we need. How do we get the values?
First Attempt: Optimization
The idea here is to do this:
For each chromosome, create an array with bp values from 0 to the length of the chromosome by 100,000. Assign a cM value to each base pair of 0.1 cM for each 100,000.
Now we take each of the matches in our segment match file and compare the actual with the cM value calculated in this table and we square the difference.
We sum the Diff Sq column. And our optimization goal is to minimum the total sum of squares by changing the cM values assigned to the 100000 bp values.
In Excel, I used their Solver tool, setting the objective as the Min of the total sum of squares cell, by allowing the algorithm to change any of the cM cells except the 0 cell. What I got was this:
Excel only allows 200 variable cells.
If you try 200 at once, it takes forever. If you try about 20 at once, it can solve the problem in a few minutes but gives some cM values lower than the previous one which is not possible. So then you have to add constraints to prevent this from happening.
This isn’t the best sort of problem for Excel to solve. Better would be to use a statistical package like R or to custom program the optimization.
Second Attempt – Following the Segment Trail
So then I thought I’d try a different tack.
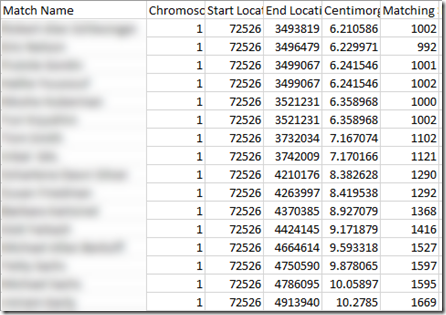
How about starting with the first match on the chromosome. For me at Family Tree DNA, on chromosome 1 that is a match from the base pair starting location 72,526. I have segments that match with 16 different people starting at that location, and they end at various locations from 3,493,819 to 4,932,655.and those segments end from 6.210586 cM to 10.2785 cM
Base pair 3,493,819 therefore is at a genomic position 6.210586 cM higher than base pair 72,526.
If for each of those end locations, I find other segments that start at that location, then I can add those segment lengths to 6.210586 to get the genomic position of the ending base pair location.
And also for all of those end locations, I can find other segments not starting at 72,526 that end at one of them, then I can subtract those segment lengths from 6.210586 to get the genomic position of the starting base pair location.
I can continue this with each base pair that is assigned a genomic position until it runs out.
I tried this for Family Tree Data. I took 10 segment match files and combined them together. I extracted Chromosome 1 and sorted by start location and end location. I eliminated duplicates because for the same start and end base pairs, the cM was always the same. That gave me 65,627 segments that covered positions 72,526 to 249,222,527.
Those 65,627 segments had 131,254 start and end positions. There were 38,225 unique positions, so each unique position was used on average in 3.4 segments.
I assigned base pair location 72,526 the genomic position 0. With the 10 files I had 21 unique segments starting at that position, compared to the 16 just for me that I show above which had 14 unique. I assigned the 21 genomic positions to the end points.
From those 21 end points, the file had 22 segments that started at one of them and 12 segments that ended at one of them that I hadn’t encountered already.
I assigned the new genomic positions to the other ends of those segments, and now I had 92 new starting segments and 177 new ending segments to process.
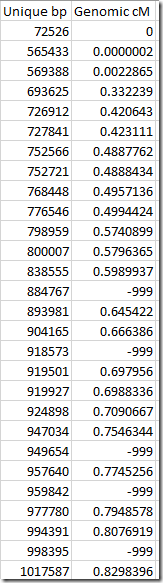
It took 20 iterations of this procedure until I ran out of segments to process. By then, I had assigned genome positions to 35,082 or 92% of the unique positions. Here is what the first few final assignments looked like:
The –999 values are those that were not assigned. If we remove those, we then have a very accurate table that can be used for determining cM length from a start base pair and end base pair for Family Tree DNA data.
Compare this to the first table in the “What is the Goal” section above. That table was not accurate enough for Family Tree DNA and you can see that the genomic position at base pair 957,898 was 2.6 cM when for Family Tree DNA, it should have been closer to 0.8 cM.
Unfortunately, I couldn’t get this method to work at 23andMe because I had a lot fewer segments to work with due tof their 1,500 person limit, and I only had 5 files from other people to combine mine with. For chromosome 1, I only had 1801 segments to work with and could only chain 76 of them together. More data would be needed for this technique to work at 23andMe.
At MyHeritage and GEDmatch, the problem is that the cM values for the segments are only given to 1 decimal point. That means each value only has an accuracy of +/- 0.05 cM. And the successive adding and subtracting of these for 20 iterations will multiply the error.
Conclusion
Well that was fun, but solving this problem is not my main goal in life. I think for now I’ll just leave it here so that someone else who gets the urge, will have some ideas to try.

Joined: Sat, 18 Mar 2023
1 blog comment, 0 forum posts
Posted: Sat, 18 Mar 2023
As always, nice writeup and approach. I have 10+ 23andMe accounts that could be used to finish your analysis (if we can reach an agreement on privacy and such :)