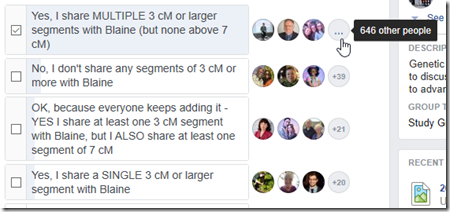
Blaine Bettinger posted a poll on his Genetic Genealogy Tips and Techniques Facebook group about 7 hours ago. It is a closed group, but if you are a member you can see the poll here. In four hours, the poll got almost 800 responses and over 350 comments.
Blaine asked people to go to GEDmatch Genesis and do a one-to-one comparison between their kit and his kit, but reduce the minimum segment threshold down from the default of 7 cM and do the comparison using a minimum of 3 cM instead.
This little poll/experiment is well designed to help people realize that most small single matching segments are false, and to realize how many of them they might have with someone whom they are likely not at all related to. It is because we have a pair of each chromosome, and there are high probabilities that alleles located on either of our chromosomes may match either of another person’s two alleles at the same position. What ends up happening is that we can get random matches to segments as large as 15 cM. As a segment gets larger, the laws of probability start saying that the random allele matches will get less likely the same way you can’t keep throwing a coin on heads forever. And above 15 cM you can be fairly certain that almost all segment matches are real, i.e. likely Identical by Descent (IBD) and likely passed down from a common ancestor that was anywhere from 1 to 20 generations back.
So here’s the result of the poll when I snapshot it:
649 out of the 738 people (88%) who responded, including me, had multiple segment matches with Blaine that were 3 cM or more but not larger than 7 cM. Only 42 people (6%) did not share any segments 3 cM or more with Blaine.
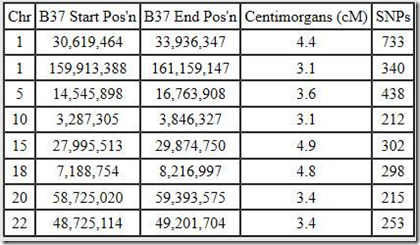
These, for example, are the segments I share with Blaine:
There’s 8 matching segments totaling just 30.7 cM. The largest is just 4.9 cM. The most SNPs shared is 733. That sort of means we have 733 SNPs in a row where one of my alleles matches one of Blaine’s alleles, but due to misreads, GEDmatch and other DNA companies usually allow for a mismatch every now and then, maybe one or two every 100 or so. Also there are usually a few percent no-calls (unreadable SNPs) that are always treated as a match. Maybe I have 15 no-calls and Blaine has 15 over those 733 SNPs, so that’s 30 positions that may not be matches but are treated so.
Blaine is rightfully trying to get the attention of genealogists to inform them to be wary of these small segment matches. They are single matches. They are dangerous, because most are false. Confirmation bias, where you think someone is related and then believe that some small segments are the connection must be avoided. I likely share zero DNA with Blaine, yet I’ve got 7 segments showing here. Don’t believe it. You need more than this.
So I thought I’d look into these small segment matches people have with Blaine and see if I could learn more. Among the 350 comments to the poll in the first 4 hours, there were 25 people who posted their matches to Blaine like I did above. I entered their matches as well as mine into a spreadsheet so I could do some analysis.
The 26 of us have 247 segment matches with Blaine. That’s on average 9.5 matches. The fewest is 2. The most is 17. The average each of us match with Blaine 35 cM, minimum 8 cM, maximum 64 cM.
If all those small segments are real, then potentially 64 cM could indicate a 3rd cousin with Blaine, but that’s the best that could happen. It is much more likely none of the 26 of us and Blaine are truly DNA related because almost all those small segment matches are false.
Does Triangulation Help?
I don’t know of any studies of triangulation done with small segments. The best I can quote is Jim Bartlett’s observations that just about all his triangulations are true down to 7 cM and most are true down to 5 cM.
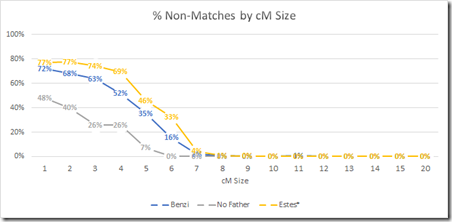
I have done a related study. It is not triangulation per se, but it is what I would call Parental Filtering. It finds segments that a child matches but neither parent matches, thus indicating that the segment match of the child is false. They cannot be matching through one chromosome (or they would match one of their parents), so they must be matching through both their chromosomes randomly. Parental filtering effectively forces the match through one chromosome just as triangulation does, so it is a good first cut estimate of how triangulation might work on small segments. The final result of my study was this graph:
This tells us that parental filtered segments almost always match when they are at least 7 cM. But between 3 cM and 7 cM, you still can have a lot of false matches even if both the child and a parent match.
So my question is, do any of the 247 segment matches of the 26 people with Blaine triangulate? If they do, is it a true match that is a small segment that is IBD, or is the triangulation a false match?
I sorted the 247 segment matches by chromosome, starting position and ending position. And I looked for overlapping segments. Guess how many there were? Would you believe 147 (60%) overlapped with one or more other segment matches. This happens because once you start to get as many as 247 segment matches, it becomes like the birthday problem (How many people in the room before 2 have the same birthday). A 4 cM match has a 1 / 500 chance of overlapping with another 4 cM match. Once you have 247 segment matches, you have 247 x 246 / 2 = 30,381 possible match combinations, so there is an expected value of 30,381 / 500 = 60 matches if they happened at random. But as the segments matches start to fill up the chromosome, the chance of matching starts to increase. So my observed number of 147 matches is quite likely. If I had entered 1000 segment matches with Blaine, almost all of them likely would overlap with at least one other segment match.
This is what I might call the “chromosome browser phenomena”. People see their segment matches lining up in the chromosome browser and assume they must be valid matches because they are all lining up. False, false false!
The chromosome browser shows you double matches. A double match is where Person A matches Person B and Person A also matches Person C on the same segment. Double matches are simply alignment of single matches. There is nothing there that tells you that any of them are valid if they are small segments.
The important step to validate a double match is to see if that match triangulates. What you need to do is check that Person B also matches Person C on the same segment. What that will usually do is, like parental filtering, force the match to be on just one chromosome between the three sets of people: A and B, A and C, B and C. (There are special case exceptions, but I won’t get into that here).
GEDmatch allows you to check triangulations. You can do a one-to-one comparison of any two people if you know their Kit number. In Blaine’s comments, about 10 of the 26 people who gave their matches inadvertently included their Kit number in their screen shot. Now I’m not going to hack their account, but I am going to do some one-to-one comparisons between them to check to see if any of these false matches are triangulations.
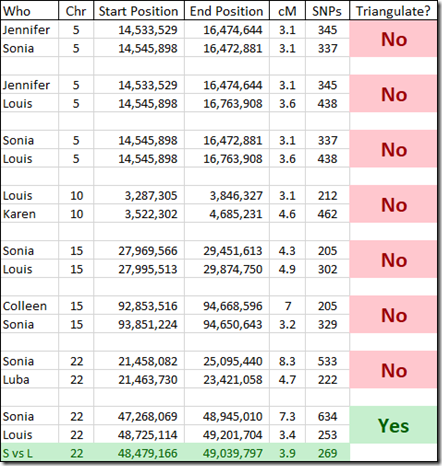
There only happened to be 8 overlapping matches for the people who have given their Kit numbers. That should do for a very rough estimate as to how many of these small segments triangulate. These are the overlaps I found and checked:
Interestingly I am in 5 of these 8 matches. I suspect the reason why I am in so many is because my kit I uploaded is a combination of the raw DNA from 5 companies. Therefore, my kit has SNPs to match all the companies, and I’m guessing that must allow me to match more segments than a single company kit would.
Checking the 8 sets of matches, I find only one of them triangulates. I’m showing the Sonia versus Louis segment match on the last line of the above table in green.
What does this all mean?
Well 7 of these 8 small segment matches do not triangulate. If a segment does not triangulate, it cannot be IBD and therefore is almost assuredly false.
This does say that triangulation a good way of further filtering out small false segments. If triangulation can eliminate 7 out of 8 segments for you, then it will have saved you a lot of unnecessary analysis.
And that 8th segment: Is it real? If the segment is above 7 cM, then Jim Bartlett would tell you that it is very likely real. But if it’s under 7 cM, then we can’t say for sure. Triangulation alone cannot prove that a small segment is real. It can only disprove a segment. You would need to analyze other matches on the same segment and check that they all match each other. Then you have a triangulation group which is genetic genealogy’s version of “a preponderance of evidence” which starts to tell you something. But there are still a lot of caveats. With small segments, each of the 3 matches may be matching randomly, or two may be matching and the third is a random match. And you may have a valid triangulation group for some people in the group, with the others in the group matching randomly to the group’s common segment.
Also note that even if you find a true triangulation group for a small segment, you must realize that a segment that small could very easily have come from an ancestor 10, 20 or even 30 generations back, so you may never find the common true ancestor for most of your small segment matches.