Writing Freeware (Double Match Triangulator) - Sun, 17 Jul 2016
Most people might think releasing a freeware program is easy. Just write it and make it available. Right?
Well, there’s a bit more in it than that.
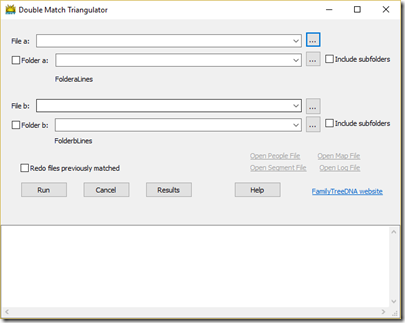
When I came up with the idea for Double Match Triangulation of autosomal DNA using the chromosome match files produced by FamilyTreeDNA, I knew I’d need a program to sort all that data out. And when I went online to see what there was, and there was nothing like it, I knew I’d have to create it and make it available so that others can use it too.
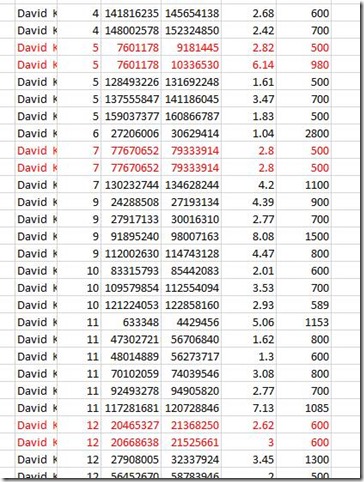
I first figured out what was needed by doing the matching with Excel. I loaded two chromosome match files into Excel, merged them together, and developed equations to determine segment overlaps. I then used conditional formatting to color the cells to make interpretation easier.
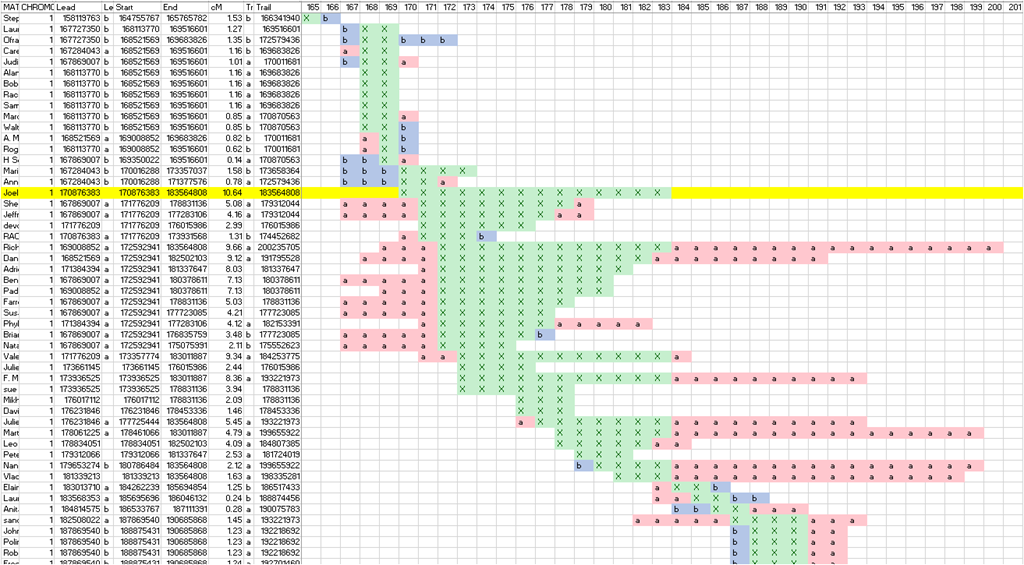
Once that template was set up, it wasn’t too much work to build a program with an engine that would read in two chromosome match files, compare them the same way I was doing in the Excel spreadsheet, and output the results to a csv (comma delimited) file so that Excel could read it in and display it all nicely.
So at that point, just a few little things to do:
2. Get a few sample files from people so I can test it.
3. Test it, and find problems with the input files and handle them.
4. Learn from the results, and figure out more that can be done.
5. Decide what will be in the first cut of the program.
Basically the program is done…. Except it’s not.
6. Mock up a user interface to allow selection of files.
7. Include Open File dialogs to select the files
8. Include Open Folder dialogs to select the folders. Wait, there aren’t any Open Folder dialogs available in the Visual Controls Library. I have to research my options, see what I did in GEDCOM File Finder, and decide how to implement this.
9. Save past files and directories to the Registry so that they can appear in the recently used list. (You’d hate me and I’d hate myself if I didn’t do this.)
10. Add error checking of file names and input files.
11. Figure out what to put in the status box and log files to track what was done and what wasn’t and any errors encountered.
12. Realize it’s easy to export to csv, but a pain to manually format it once you load the csv file into Excel. So I look for a way to automate the loading of the Excel file directly.
13. Try to make sense of the Office Developer Documentation and find the commands needed amongst the millions of articles.
14. Spend a week implementing the automation, and once it is working, realize it takes 10 times longer than creating the csv file.
15. Puzzle about ways to improve this slowness while in the shower, on my bike and at 3 in the morning.
16. Try various things, and find that creating a temporary csv file and then automating its input is 5 times faster than direct to Excel automation.
17. Rewrite everything so that multiple files can be matched at once.
18. Make sure it all looks nice, still works, and does what’s needed.
All done now? Yup. Except left to do:
19. An installation script for it.
20. Webpage for it so there’s someplace to download it from.
21. Some documentation would be nice.
22. Blog posts, announcements
Yay! Finally done. … but forever followed by:
23. Support, bug fixes, response to questions, enhancements
So that’s how a freeware program is made. And the timeframe is after work, in the evenings and on weekends when not on errands, when your family lets you be alone, and when you’re not too tired to think.
Hopefully the Double Match Triangulator program will be available in the next week or two for anyone to try out.



 Feedspot 100 Best Genealogy Blogs
Feedspot 100 Best Genealogy Blogs





