I’ve released a beta of what will be version 1.2 of DMT.
It has all the new functionality. I just haven’t updated the help file yet. Once the help file is updated, I’ll re-release it as 1.2.
With the deadline for the RootsTech Innovator Showdown coming up, I wanted to get some major enhancements in. There are a number of exciting changes.
1. Double Match and Triangulation Groups
DMT now will group the matching segments together based on the crossovers between them. Each group will be delineated by a thick box around them.
The idea came from the two new posts by Jim Bartlett on his Segmentology blog. Understanding and Using TGs followed the next day by The Attributes of a TG. In the latter article in section A6, Jim explains how he identifies crossover points and the 6 steps to to find the end location of his Triangulation Groups.
I thought about this, and worked to implement something similar. Believe me when I say I must have changed the procedure a dozen times from my original idea before I got it working. I had to translate what Jim was saying to the data structures of my program.
Along the way, I learned a lot, including the following:
- Jim works with single matches that he is manually triangulating. That’s an admirable mass of work and he’s successfully mapped the majority of his segments. He talks about the ends being fuzzy. That is true for single matches because there are often some random matches at the ends. Although I have no proof, my observations indicate that the double match ends seem to be mostly precise. The random ends just don’t seem to be there. Crossover base addresses look very precise. All the single matches with fuzzy ends are excluded.
- I realized from Jim’s work is that the goal is to make a map of all the ancestors of Person a. You need the b people to do the double matching, but the goal is to map out a single person. Therefore, it is only the green X’s (double matches) and red a’s that should be used to define the Triangulation Groups.
- I needed an algorithmic way to determine the start and end of each Triangulation Group. My DMT program couldn’t use “judgement” because there’s “no hard rule”. So I developed a rule, the rule being: If the next segment starts at or after the end of the previous segment, then end the last Triangulation Group and start the next Triangulation Group.
- To make that rule work, I had to sort the segments first by the lowest starting base address, and then by the highest ending base address. You can see in the diagram above, that the triangulation groups get smaller as you go down the page, and then when the start point changes, they start again. Larger TGs are closer ancestors that are made up of smaller TG’s that are segments from ancestors of those ancestors.
- I have learned that Double Match segments that don’t Triangulate are as valuable as Double Match segments that do Triangulate. I call them Missing a-b Matches and I wrote about them in an earlier blog post. Triangulated segments belong to a common ancestor. Missing a-b matches may match from both halves of an ancestor’s chromosome, or may match from the same segment address of two ancestral parents (a father and mother). The Double match means that Person a matches Person c and Person b matches Person c. The likelihood of two matches to the same person, both by chance is severely reduced, especially since these are people in common match lists. I believe Jim Bartlett’s suggestion that Triangulated segments are mostly IBD down to 5 cM likely also applies to Double Match groups without the a-b match.
Once I got this going, I saw that I could improve the information shown in the Map file. In the above diagram, each row is one double match. The columns are:
- Person a – the main person you are matching to
- Person b – the person you are double matching with
- Person c – the person who double matches both a and b on the segment
- Chr – the chromosome number. 23 = the X chromosome.
- Start-AC – the base address where the a-c match starts
- End-AC – the base address where the a-c match ends
- CM-AC – the distance the a-c match covers in cM
- Start-BC – the base address where the b-c match starts
- End-BC – the base address where the b-c match ends
- CM-BC – the distance the b-c match covers in cM
- DMG-START – the base address where the Double Match Group starts
- DMG-END – the base address where the Double Match Group ends
- DMGROUP – Jim Bartlett’s name for the Double Match Group
- STATUS – tells you if that segment triangulates
Once again, Double Match Groups (DMG) are segments where a number of people have double matches and indicate common ancestors. If a Double Match Group also has the a-b match, then it s also a Triangulation Group (TG) and indicates a common ancestral segment on the same half of the chromosome.
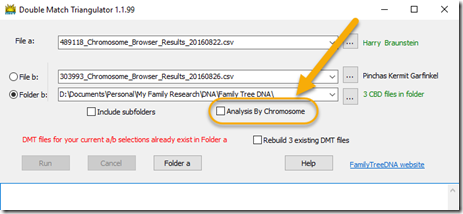
2. Analysis By Chromosome
So that was the major improvement for version 1.2 of DMT. But that’s not all. I’ve added another checkbox to the main window:
If you check the Analysis By Chromosome box, then DMT will combine every individual run between Person a and all the b people that are in Folder b. It will then place the results into 23 files, one for each Chromosome.
The main reason why the chromosomes are split up is because the files can become very large. In my case, I have the Chromosome Browser Download files from 61 related people that I combine with my uncle’s. It takes DMT about 5 minutes to process all the files and produce the 23 chromosome map files. It’s an amazing amount of information that I believe may unlock all the secrets we’re looking for … if we can figure out how to do so. I’ll be trying. And if you figure something out, let me know, and maybe I can program it in for everyone to use.
For example, I’m sure there is enough information in the crossover base addresses for DMT to split the Double Match Groups into the two sides: maternal and paternal. That will be something I’ll be looking at to figure out how to do for version 1.3 and I’ll likely call it Double Match Filtering.
3. Improved People Page/File
Along with the 23 map files the Analysis By Chromosome produces, a combined People file is also produced. It looks like this:
This file will help you locate the relevant segments for the people you are interested in, and determine which chromosome file they’ll be in. All of Person a’s Person c matches are listed by highest total cM. The maximum segment length of each a-c match for each Chromosome is shown. That is followed by one column for every Person b so you can determine all the people Person c double matches to.
Like I say, I’m still trying to figure all this out myself. But feel free to try it out.
If you notice any problems or have any ideas, please let me know.
—-
Note: Version 1,1.99.1 corrects a problem where not all Triangulated segments were identified. If you downloaded 1.1.99 between October 18 and October 20, please download 1.1.99.1 from the link at the top of this post).